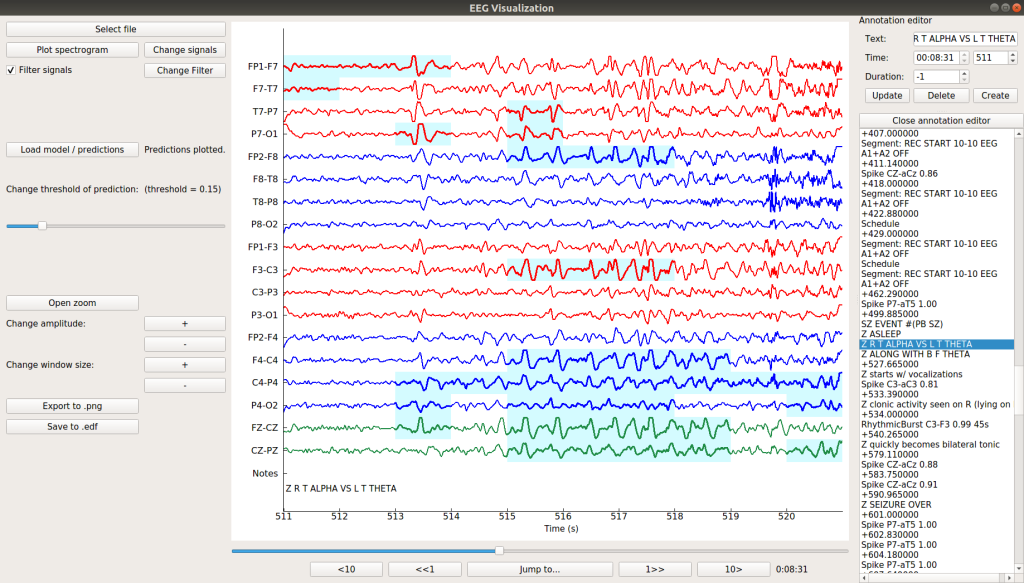
EPViz (EEG Prediction Visualizer)
Scalp EEG is one of the most popular noninvasive modalities for studying real-time neural phenomena. While traditional EEG studies have focused on identifying group-level statistical effects, the rise of machine learning has prompted a shift in the community towards spatio-temporal predictive analyses. We have developed the EEG Prediction Visualizer (EPViz) to aid researchers in developing, validating, and reporting their predictive modeling outputs. EPViz is a lightweight and standalone software package developed in Python. Beyond viewing and manipulating the EEG data, EPViz allows researchers to load a PyTorch deep learning model, apply it to the EEG, and overlay the output channel-wise or subject-level temporal predictions on top of the original time series.
These results can be saved as high-resolution images for use in manuscripts and presentations. There is also a command-line option for batch processing. EPViz also provides valuable tools for clinician-scientists, including spectrum visualization, computation of basic statistics, data anonymization, and annotation editing.
EPViz can be installed in three ways: (1) cloning our GitHub repository to access the latest version, (2) through PyPI, and (3) as a standalone prepackaged application for MacOS and Windows. [github][pypi][Mac App][Windows App].
Please visit our EPViz page for the complete user guide.
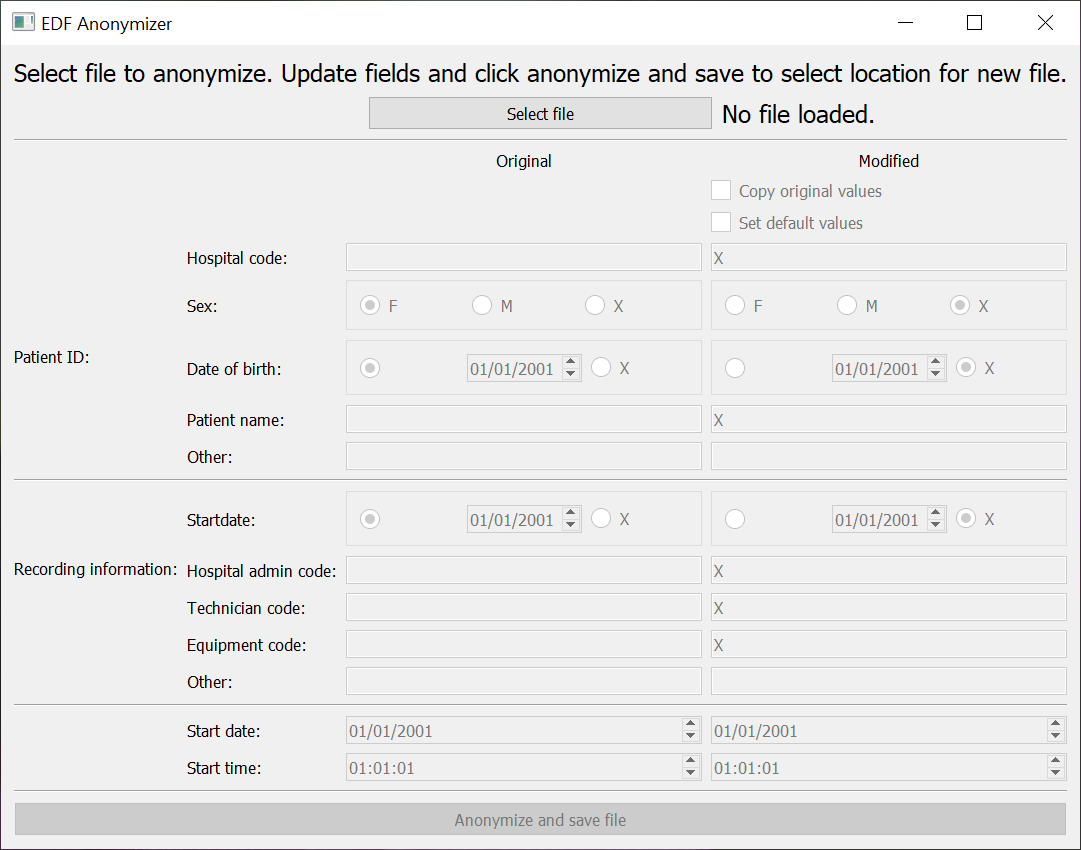
EDF Anonymizer
We have also provided just the anonymization tool in EPViz as a standalone module. This tool allows the user to alter the EDF header fields and provides default settings for scrubbing patient IDs and time stamps. [github][Windows App][MAC App]
Epilepsy
- DeepSOZ: A Graph Convolutional Network for Automated Seizure Onset Zone Localization from Resting-State fMRI Connectivity.
N. Nandakumar, D. Hsu, R. Ahmed, A. Venkataraman.
Under Revision for IEEE Transactions on Biomedical Engineering, 2022. [paper][github]
Predictive Connectomics
- A Matrix Auto-encoder Framework to Align the Functional and Structural Connectivity Manifolds as Guided by Behavioral Phenotypes.
N.S. D’Souza, M.B. Nebel, D. Crocetti, N. Wymbs, J. Robinson, S. Mostofsky, A. Venkataraman.
In Proc. MICCAI: Medical Image Computing and Computer Assisted Intervention, LNCS (to appear), 2021. [paper][github] - M-GCN: A Multimodal Graph Convolutional Network to Integrate Functional and Structural Connectomics Data to Predict Multidimensional Phenotypic Characterizations.
N.S. D’Souza, M.B. Nebel, D. Crocetti, N. Wymbs, J. Robinson, S. Mostofsky, A. Venkataraman.
In Proc. MIDL: Medical Imaging with Deep Learning, MLR:1-12, 2021. [paper][github] - A Deep-Generative Hybrid Model to Integrate Multimodal and Dynamic Connectivity for Predicting Spectrum-Level Deficits in Autism.
N.S. D’Souza, M.B. Nebel, D. Crocetti, N. Wymbs, J. Robinson, S. Mostofsky, A. Venkataraman.
In Proc. MICCAI: Medical Image Computing and Computer Assisted Intervention, LNCS 12267:437-447, 2020. [paper][github] - A Joint Network Optimization Framework to Predict Clinical Severity from Resting State fMRI Data.
N.S. D’Souza, N. Wymbs, M.B. Nebel, S. Mostofsky, A. Venkataraman.
NeuroImage, 206:116314, 2020. [paper][github]
Preoperative Mapping with Resting-State fMRI
- Multi-Scale Spatial and Temporal Attention Network on Dynamic Connectivity to Localize The Eloquent Cortex in Brain Tumor Patients [paper][github]
- A Multi-Task Deep Learning Framework to Localize the Eloquent Cortex in Brain Tumor Patients using Both Static and Dynamic Functional Conn [paper][github]
Emotional Speech
- Sample, Attend and Morph: A Deep-Bayesian Framework for Adaptive Speech Duration Modification [paper][github]
- Non-parallel Emotion Conversion using a Deep-Generative Hybrid Network and an Adversarial Pair Discriminator [paper][github]
- Multi-Speaker Emotion Conversion via Latent Variable Regularization and a Chained Encoder-Decoder-Predictor Network [paper][github]
- A Diffeomorphic Flow-based Variational Framework for Multi-speaker Emotion Conversion [paper][github][sample audio]