Research
Epileptic Seizure Detection and Localization
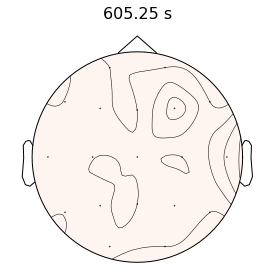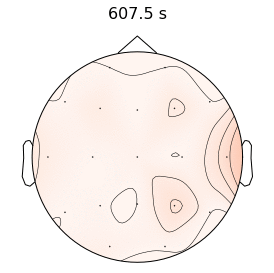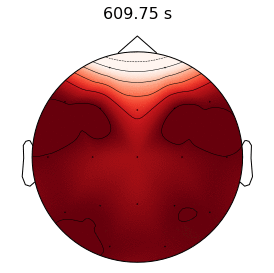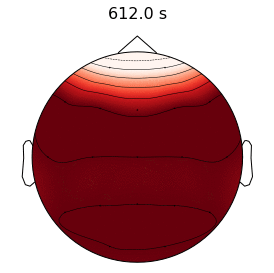
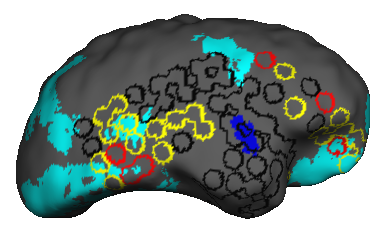
Epilepsy affects nearly 3.5 million people in the United States and is linked to a five-fold increase in mortality. While epilepsy is often controlled with medication, 20-40% of patients are medically refractory and continue to experience seizures in spite of drug therapies. If we can determine that the seizures originate from an isolated area, then an alternative treatment is to surgically resect this seizure onset zone (SOZ). Noninvasive SOZ localization remains the linchpin for effective therapeutic planning and patient care. However, it relies almost exclusively on visual inspection, which is time consuming and prone to human error. Our goal is to develop an automated platform for reliable, noninvasive SOZ localization that complements the existing clinical workflow.
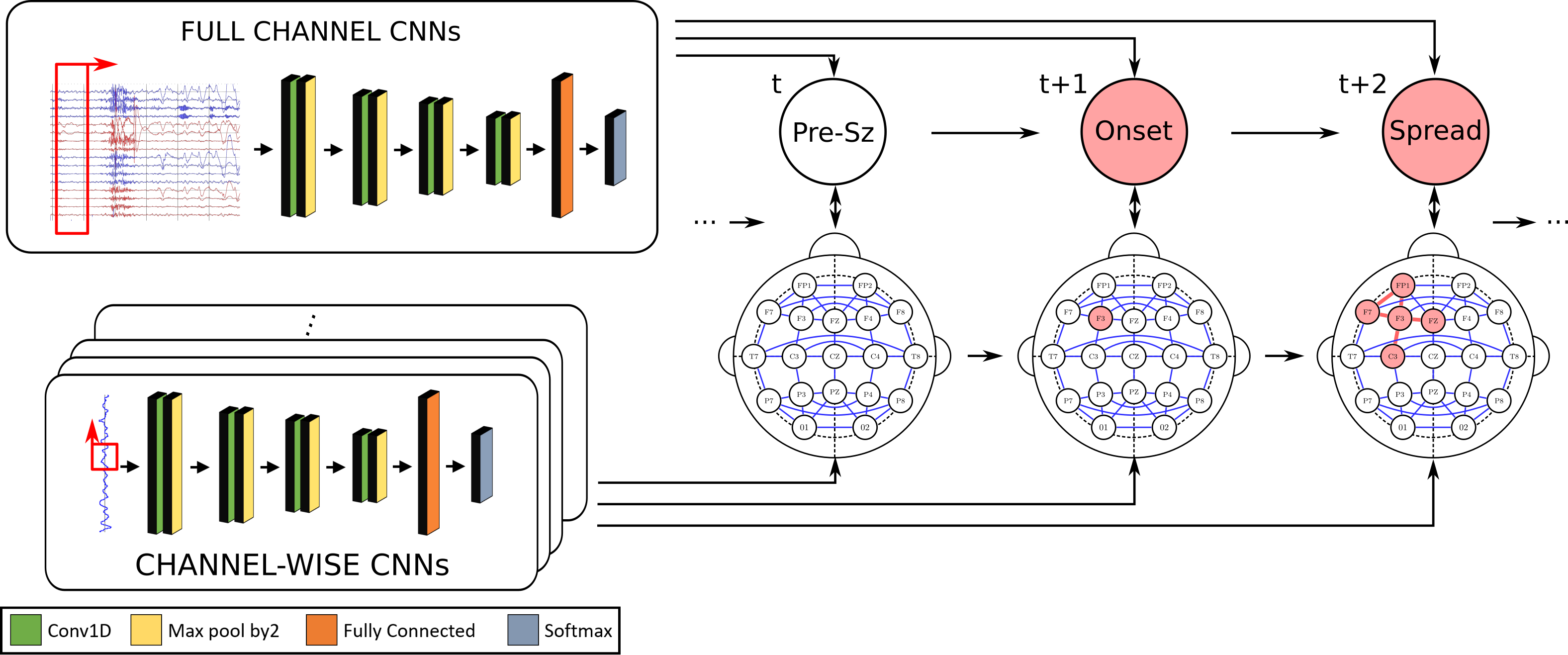
We have designed a suite of methods for automated seizure detection and tracking from scalp EEG data. These methods use probabilistic graphical models track the evolving spatiotemporal propagation of a seizure to infer the onset. We have validated our approach on data from the Johns Hopkins Hospital, the Children’s Hospital in Boston, and the University of Wisconsin Madison. Our ongoing work is focused on improving the performance and robustness of the seizure tracking models via convolutional-recurrent deep learning architectures and novel data fusion training strategies.
In parallel to scalp EEG, we are working on methods to localize the SOZ from resting-state fMRI data. The core assumption is that focal abnormalities in the brain will cause localized disruptions in the functional connectivity to/from the associated regions. We have developed both a fully-parameterized probabilistic graphical models and a data-driven graph convolutional network to solve this challenging SOZ localization task.
SELECTED PUBLICATIONS
A Biologically Interpretable Graph Convolutional Network to Link Genetic Risk Propagations and Imaging Biomarkers of Disease.
S. Ghosal, Q. Chen, G. Pergola, A.L. Goldman, W. Ulrich, D.R. Weinberger, A. Venkataraman.
To Appear in International Conference on Learning Representations (ICLR), 2022.[Acceptance Rate ≈ 30%]
A Generative Discriminative Framework that Integrates Imaging, Genetic, and Diagnosis into Coupled Low Dimensional Space.
S. Ghosal, Q. Chen, G. Pergola, A.L. Goldman, W. Ulrich, K.F. Berman, A. Rampino, G. Blasi, L. Fazio, A. Bertolino, D.R. Weinberger, V.S. Mattay, A. Venkataraman.
NeuroImage: 238:118200, 2021.
G-MIND: An End-to-End Multimodal Imaging-Genetics Framework for Biomarker Identification and Disease Classification.
S. Ghosal, Q. Chen, G. Pergola, A.L. Goldman, W. Ulrich, K.F. Berman, A. Rampino, G. Blasi, L. Fazio, A. Bertolino, D.R. Weinberger, V.S. Mattay, A. Venkataraman.
In Proc. SPIE, vol.11596, 2021. Selected an Oral Presentation (<15% of Papers) – Best Paper Award
Bridging Imaging, Genetics, and Diagnosis in a Coupled Low-Dimensional Framework.
S. Ghosal, Q. Chen, A.L. Goldman, W. Ulrich, K.F. Berman, D.R. Weinberger, V.S. Mattay, A. Venkataraman.
In Proc. MICCAI: International Conference on Medical Image Computing and Computer Assisted Intervention, LNCS 11767:647-655, 2019. [Acceptance Rate ≈ 30%]
Selected for Early Acceptance (Top 18% of Submissions)
FUNDING