RESEARCH
Our Methods
Our approach to investigate protein folding, dynamics, function and assembly is deeply interdisciplinary, combining theory and experiments, and utilizing a whole assortment of techniques, including protein engineering and design, cutting-edge biophysical experimental methods (single-molecule fluorescence microscopy, single-molecule force spectroscopy, nuclear magnetic resonance, ultrafast kinetic methods), theoretical modeling and computational simulations. Graduate students and postdocs working in the lab specialize in some of these approaches towards particular project goals, or work in teams with our group members and/or some of our external collaborators for those projects that require a broader palette of techniques and a more holistic approach.
Protein Engineering and Design
We work with proteins, studying their behavior and/or trying to make them better. Therefore, a very important component of our research involves exploiting the tools of genetic engineering (cloning, DNA sequencing, site-directed mutagenesis, PCR, gene synthesis), recombinant protein production (bacterial culture, recombinant protein expression) and protein biochemistry (protein purification and characterization) to make and modify all the proteins that we work with. We also employ computational modeling methods for designing proteins and the type of mutations or artificial changes that we want to introduce on our target proteins to provide them with new or enhanced properties.
Biophysical and Structural Characterization of Proteins
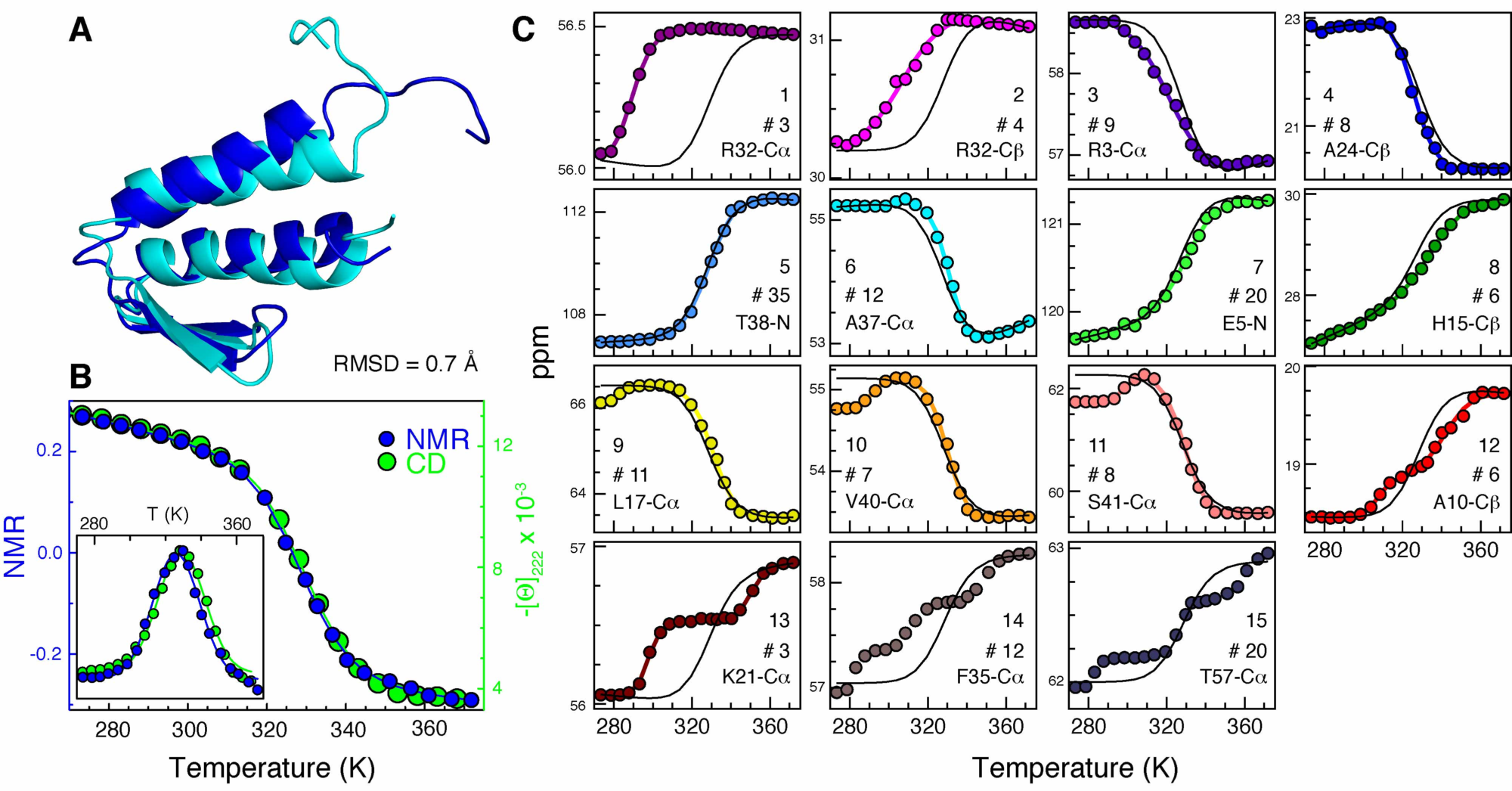
A very important step in almost any project we work on requires a detailed characterization of the conformational behavior of the protein. We look into the conformation, stability, folding and functionality of proteins using several spectroscopic methods such as fluorescence (quantum yield, spectral shifts, anisotropy, lifetime, Forster resonance energy transfer), uv circular dichroism and infrared absorption to look at protein secondary structure, and nuclear magnetic resonance combined with mass spectrometry and differential scanning calorimetry (in collaboration with the group of Jose Manuel Sanchez-Ruiz at the University of Granada, Spain).
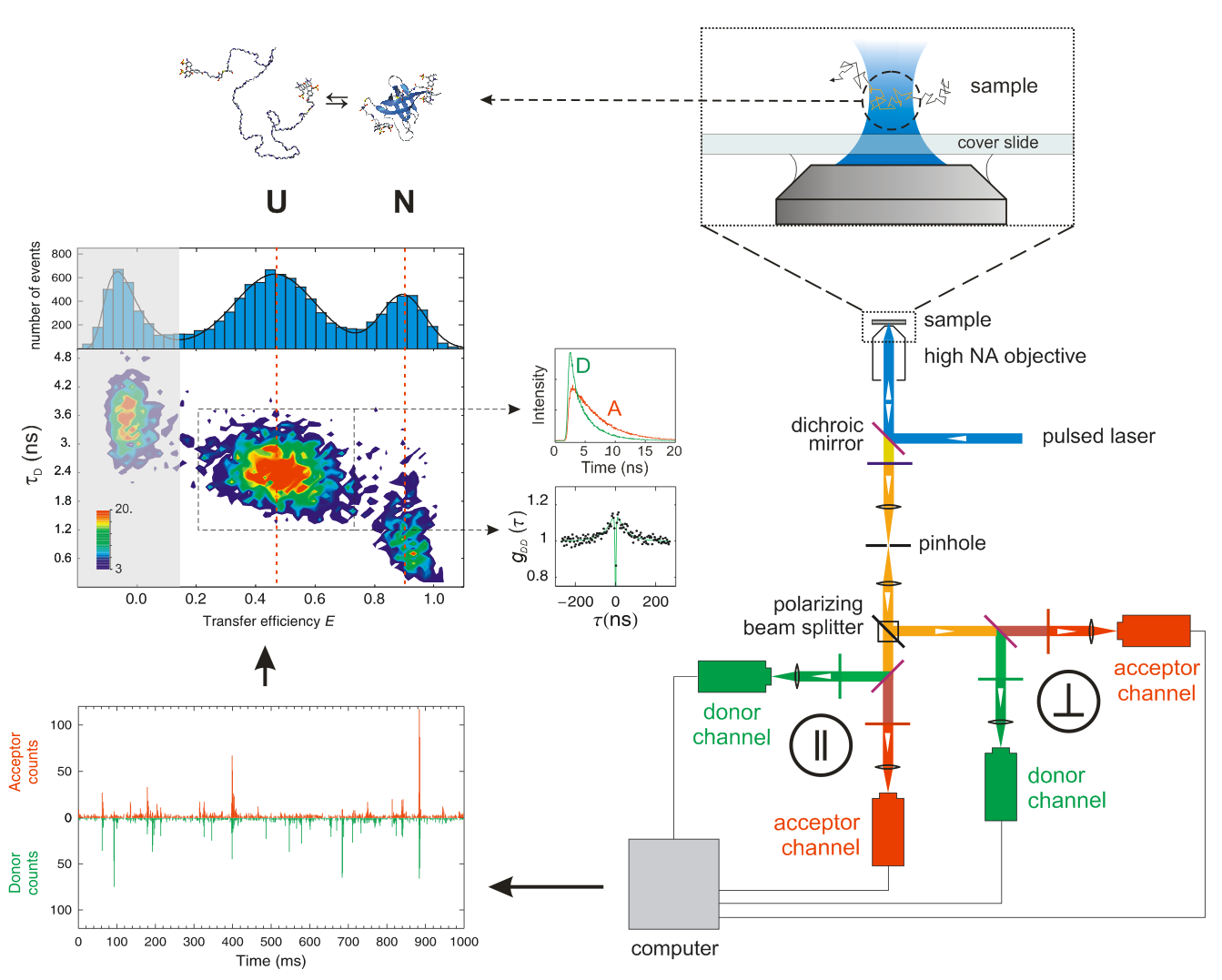
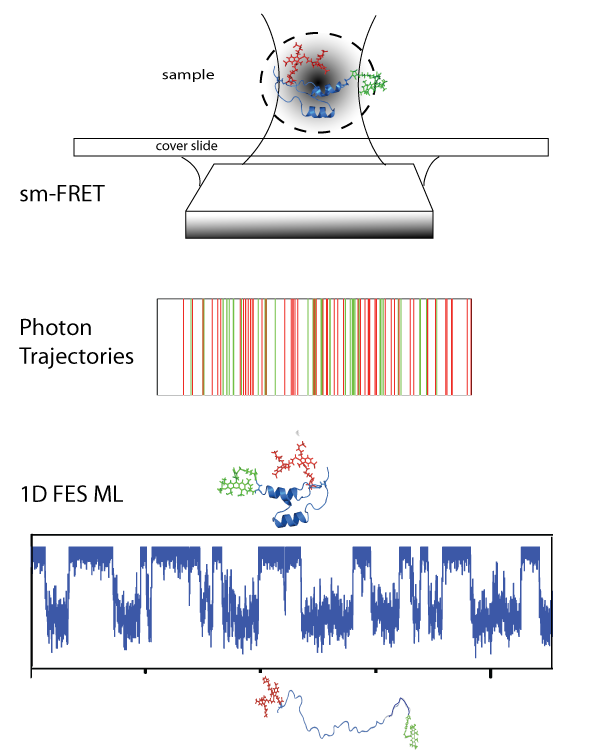
Single-Molecule Detection Methods
One of the most intriguing characteristics of proteins is that their complex conformational and functional behavior are controlled by networks of weak non-covalent interactions (intra- and inter-molecular) and driven by stochastic fluctuations induced by thermal energy. To investigate their most intimate properties it is thus essential to resolve the behavior of individual molecules while they are performing their task, whether folding and unfolding, binding to partners, looking for their target site in genomic DNA, or assembling. We use a variety of methods to monitor the fluctuations of individual protein molecules based on single-molecule fluorescence microscopy using instruments that are custom-built in our laboratory. Single-molecule confocal microscopy allow us to watch the conformational transitions of individual protein molecules with sub-microsecond time-resolution. Total internal reflection fluorescence microscopy allows us to monitor complex interactions that require a wide-field of observation and single-molecule resolution such as watching the sliding and gliding of transcription factors over DNA or the formation of large macromolecular assemblies. We use fluorescence correlation spectroscopy to measure the binding affinity of protein-protein and protein DNA interactions and the conformational dynamics of proteins. We also employ single-molecule force spectroscopy to monitor the mechanical stability of folded proteins and protein complexes as well as to exert mechanical control over specific processes. We do this in collaboration with the group of Raul Perez-Jimenez at the Nanogune Research Institute in Spain, using advanced atomic force spectrometer and magnetic tweezers. We also have plans to implement a combined magnetic tweezers and single molecule confocal fluorescence instrument that will allow us to monitor processes by fluorescence while we control them mechanically by force.
Nuclear Magnetic Resonance
We use high-resolution nuclear magnetic resonance to investigate the native structures of proteins, study their folding pathways and map the networks of non-covalent interactions that define folding mechanisms, and resolve the conformational dynamics of proteins while they are performing their job, like for example in the search of transcription factors over DNA or in the folding coupled to binding processes of intrinsically disordered proteins. The specific NMR methods that we use include multidimensional NMR methods for protein structure determination, and a variety of NMR relaxation techniques including relaxation dispersion (RD-NMR) and paramagnetic relaxation enhancement (PRE-NMR). For the multidimensional NMR work on protein folding we have a long standing collaboration with the group of Eva de Alba, a colleague at UC Merced. On the relaxation dispersion NMR experiments we have collaborated with the group of Lewis Kay at the University of Toronto and are collaborating with Nicola D’Amelio, a former postdoc of the group, now a group leader at the University of Picardie Jules Verne in Amiens, France.
Ultrafast Kinetic Methods
Our group played a pioneering role in the development of the fast folding sub-field in which ultrafast kinetic methods were used to monitor key aspects of protein folding reactions. Although the focus of our research has shifted more towards single-molecule methods, we still use fast kinetic experiments because of our interest in conformational motions, downhill folding, and folding coupled to binding which take place in microsecond timescales. Specifically, we have a custom-built microsecond continuous flow instrument based on microfluidics that we use to investigate the folding properties of downhill folding proteins.
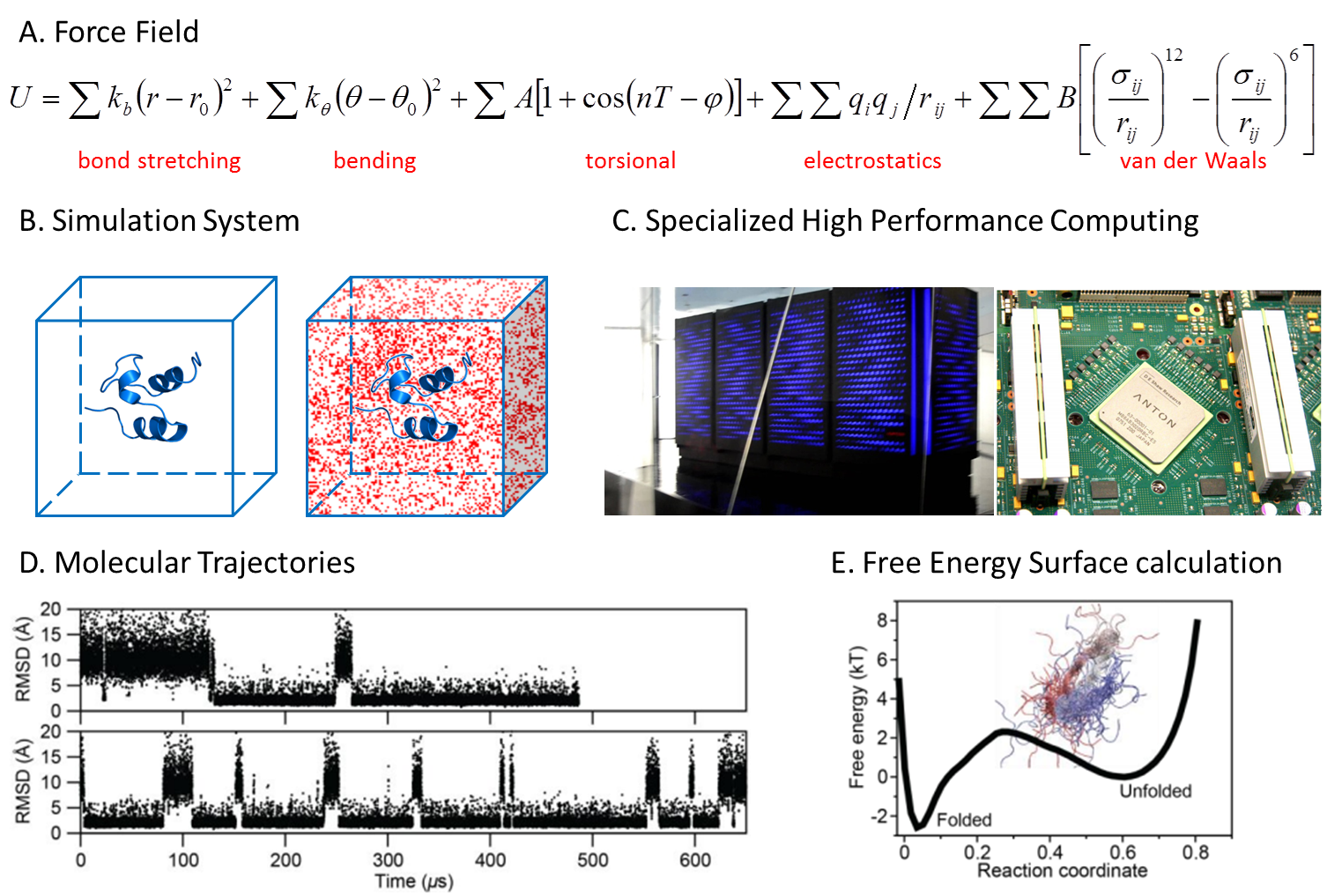
Computational Methods: Molecular Dynamics and Coarse-Grained Simulations
In conjunction with all the biophysical experiments described above, we make extensive use of theory and computer simulations. We use statistical mechanical modeling to reproduce (fit) and interpret quantitatively many of the experiments we do in the lab, as well as to make predictions that we can then test with new experiments. Computer simulations give us the opportunity to investigate the intimate behavior of conformational processes of proteins and their interactions with a higher level of structural detail and control that is attainable by experiment. Atomistic simulations are used to reproduce and rationalize experiments in truly mechanistic ways (for example steered molecular dynamics simulations vs. single-molecule mechanical unfolding experiments, or long-timescale molecular dynamics simulations vs. NMR analysis of folding interaction networks). Coarse-grained molecular simulations allow us to carefully manipulate and control the molecular properties of proteins and complexes so that we can perform targeted in-silico experiments in conditions that are not accessible to physical experiments.