RESEARCH
Molecular Aspects of Gene Expression
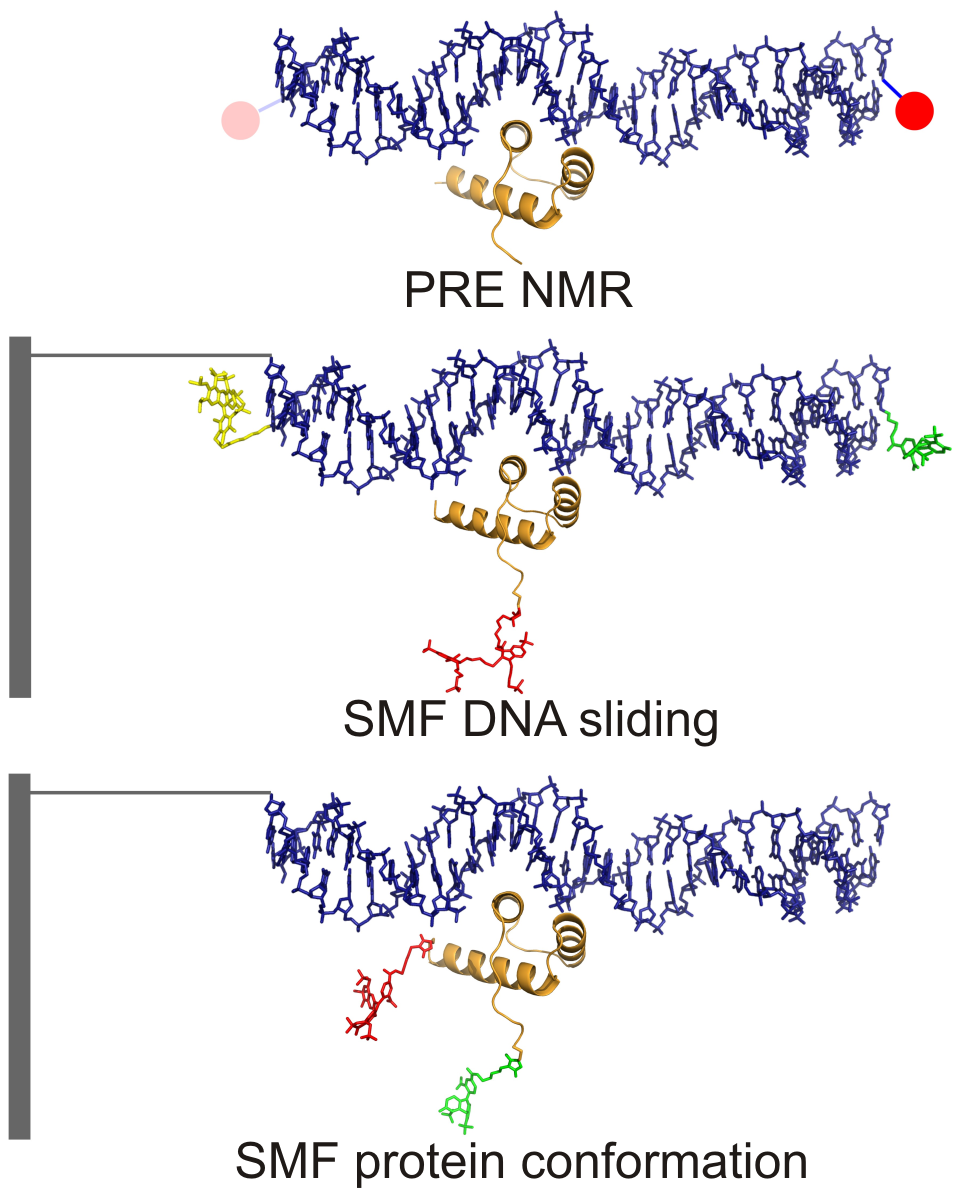
Specific DNA recognition by transcription factors is fundamental to gene expression. These DNA-binding proteins, which are present in nanoM concentrations in living cells, must efficiently find and recognize their specific target site (just a few base pairs long) among the millions of alternative nonspecific sites present in genomic DNA. An intriguing implication is that the rate by which these proteins bind to their target DNA greatly exceeds the theoretical limit imposed by the occurrence of random collisions between the protein and the DNA specific site. The way that these proteins are thought to solve that problem is by binding non-specifically to DNA and sliding while bound. The combination of sliding (1D diffusion) and hopping (3D diffusion) reduces the dimensionality of the search process and thus speeds it up. DNA sliding has been studied theoretically and computationally, and also observed experimentally using single-molecule methods and PRE-NMR.
There are, however, many questions about the search and find mechanism of transcription factors that remain open, especially from the protein viewpoint. For instance, according to the 1D diffusion model, DNA-binding domains need to implement two binding modes: high affinity to the target sequence and low-affinity to non-specific DNA sequences. But, how do they implement these two binding modes in an effective way? The model also raises an important paradox: how does the protein simultaneously maximize speed and stability? These two properties are in direct conflict because an efficient 1D search requires processivity (strong non-specific interactions), but the stronger the binding the slower the 1D diffusion coefficient since the protein will need to break strong interactions to move forward. Moreover, what are the typical lengths of effective 1D diffusion over DNA, and how do they compare with the genomic distances between genes controlled by the same transcription factor? And what is the spatial distribution of the transcription factor inside the cell?
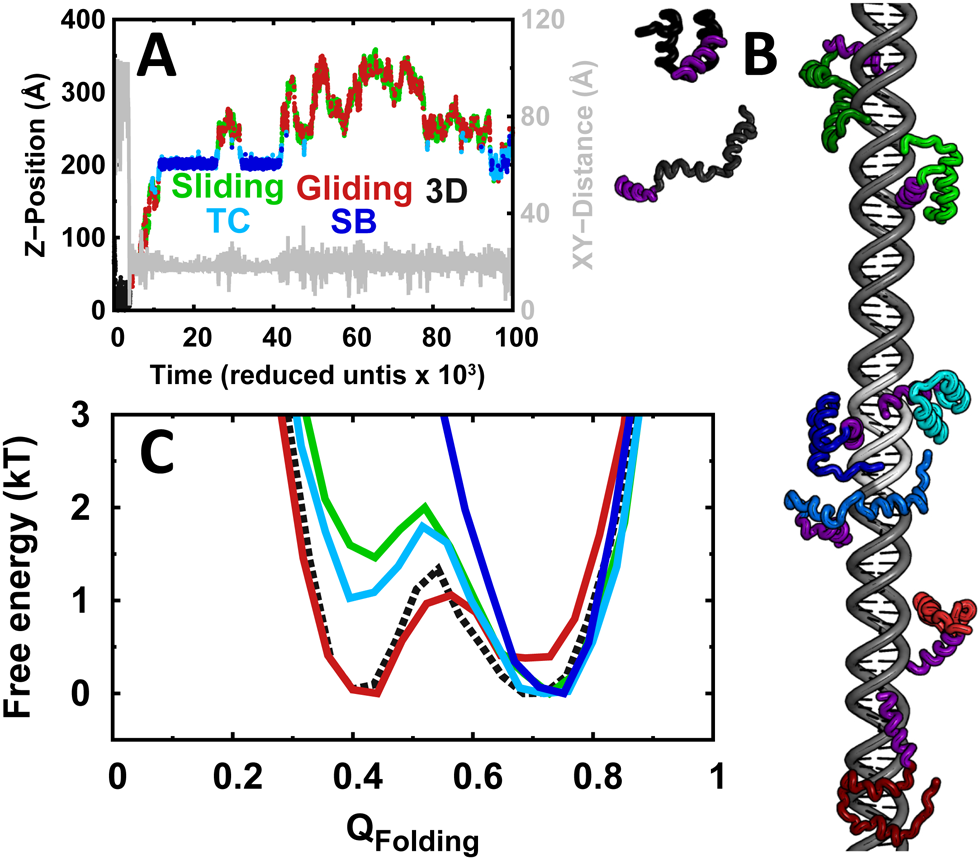
Moreover, how does the protein lock into the target site while it is sliding over DNA? Are there any homing mechanisms to guarantee that the quickly sliding protein does not overlook the target site? And if so, how are they implemented and coordinated with the search mode? We are very interested in answering all of these questions in quantitative and mechanistic terms. To investigate these issues, we take advantage of our in-depth understanding of protein folding and dynamics and the extensive toolset that is available to us and which includes advanced biophysical methods, computer simulations and protein engineering and design. Our model system to investigate these questions is the protein eng-HD, which is the ~65-residue, -helical DNA binding domain of the engrailed transcription factor from the fruit fly (D. melanogaster), and the many genes that are controlled by engrailed during development.