RESEARCH
Engineering Nanoscale Biosensors
A major challenge in biosensor research is to achieve specific sensing at nanoscale (molecular) resolutions in living cells and in real time. Proteins are in principle ideal biosensor scaffolds because of their high specificity and tunable affinity for binding, and their built-in mechanism for transducing signals through conformational changes. However, reaching nanoscale resolution in real time requires single-molecule devices that produce analog outputs. This is a serious limitation because typical proteins behave as molecular switches with inherently binary outputs: in the presence of a specific molecule to which a protein binds tightly, the unfolded protein folds to its unique correct conformation and produces a signal such as fluorescence; whereas in the absence of the ligand molecule the protein unfolds and the signal stops altogether.
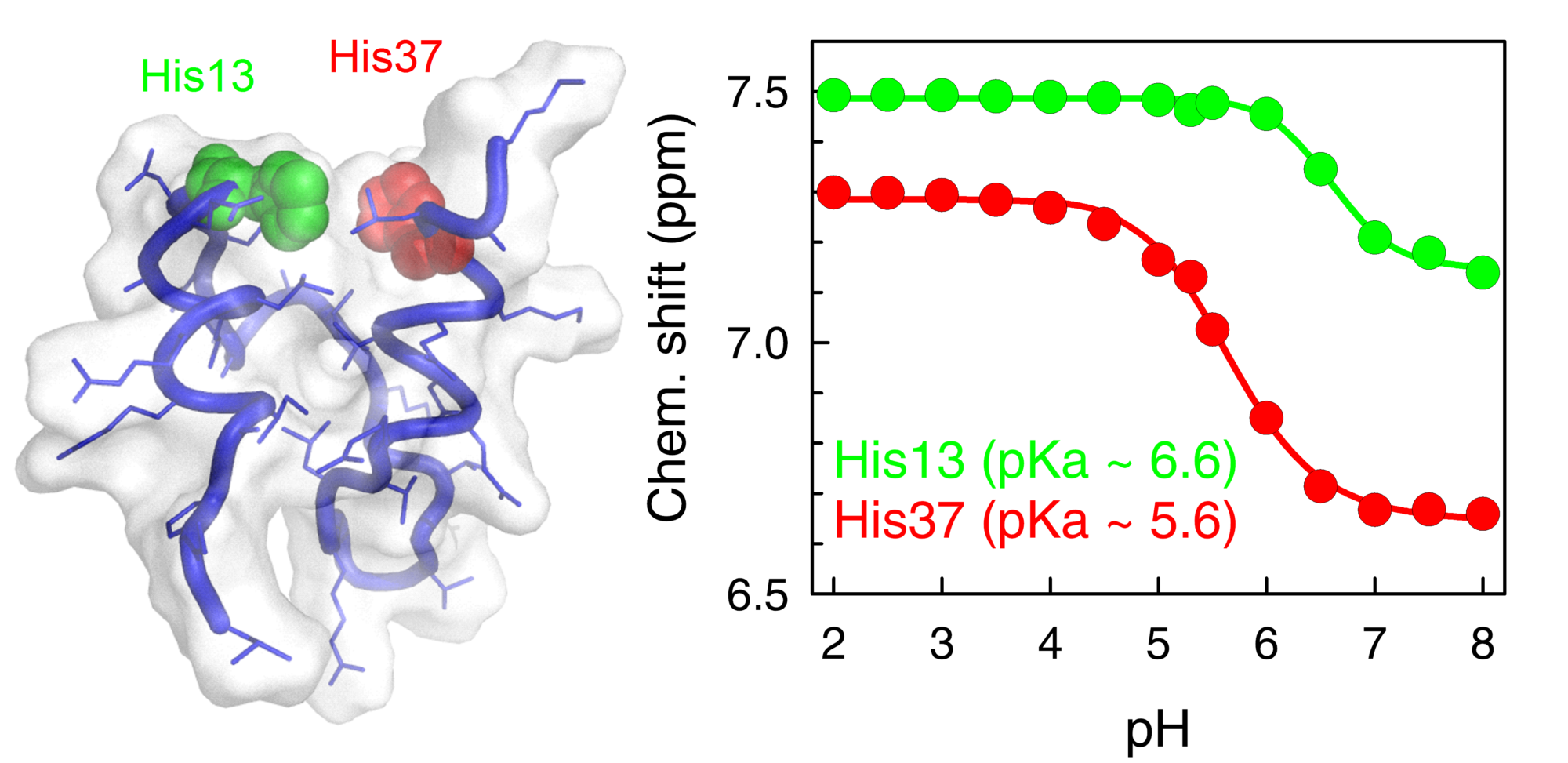
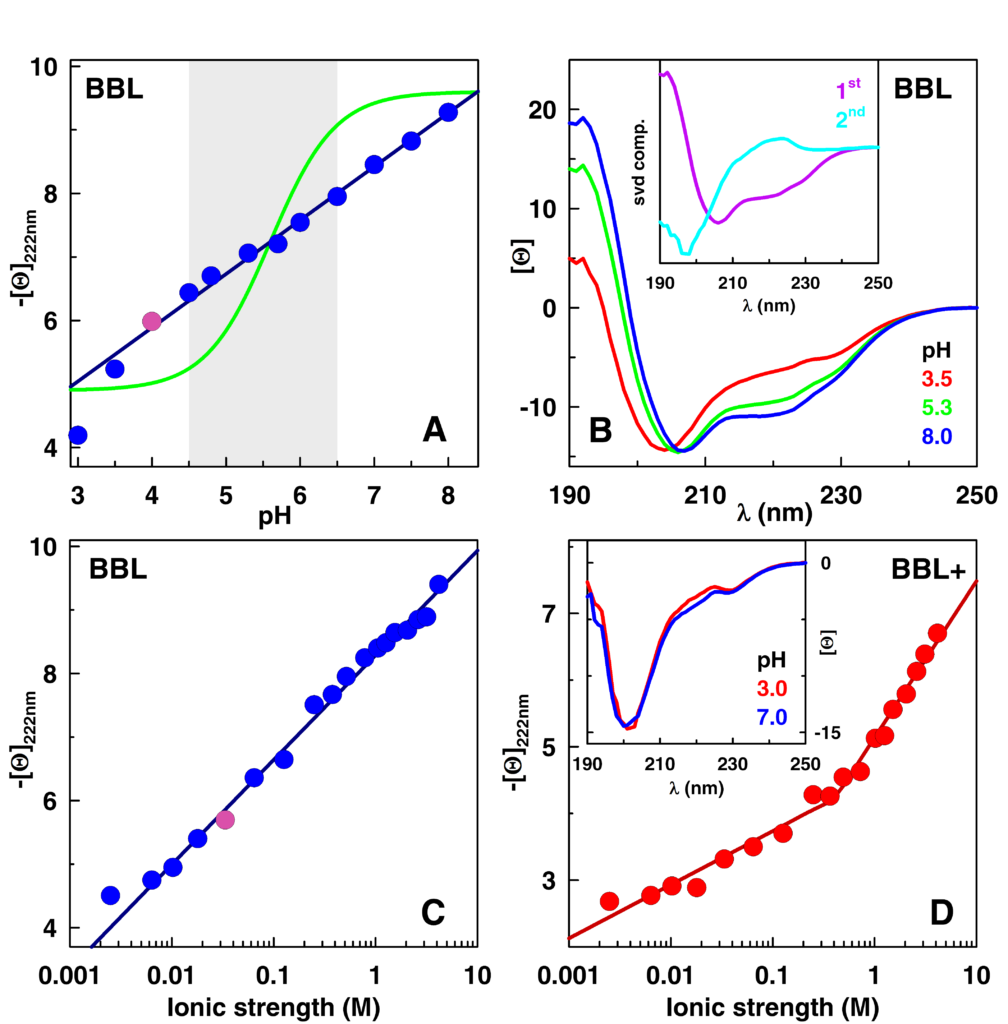
Our group is pioneering an approach to break this limitation and thus be able to develop single-molecule biosensors based on proteins. This approach capitalizes on our previous discovery of “downhill folding” proteins, i.e., proteins that fold and unfold gradually through a continuum of partially-folded intermediates not separated by free-energy barriers. Downhill folding proteins are theoretically and computationally predicted to detect other molecules in an analog mode, much like a rheostat. To prove this hypothesis and develop proof of concept analogical single-molecule biosensors we are applying the “folding coupled to binding” general biosensor-design principle to proteins purposely engineered to fold downhill. These proteins will change conformational gradually upon binding to their specific effector, creating a single-molecule analogical transducer. By introducing a high sensitivity fluorescence readout into these proteins (a fluorescence signal that is sensitive to the structural change occurring when the protein binds the analyte) we intend to attain the sensitivity required for single-molecule monitoring. These novel sensors are expected to display wide dynamic range, ultrafast response and analog readouts at the single molecule level. We have already shown that the downhill folding protein BBL can be engineered into a super-broad and ultrafast pH and ionic sensor (Cerminara JACS 2012). We have also developed a novel procedure to introduce a tunable pH transducer into any given protein by grafting histidine residues into highly buried positions within the protein core. We have a number of projects in this applied research area in which we are using these ideas and methods to try and develop single-molecule pH sensors, Ca2+ sensors, and Cu2+ sensors.