RESEARCH
Conformational Rheostats and Folding Coupled to Binding
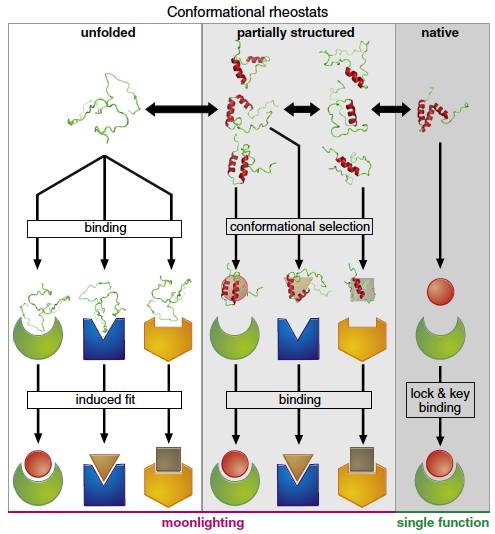
Extensive work on protein biophysics over the last two decades has unveiled a plethora of exceptions to the classical paradigm by which proteins fold into unique 3D structures in single strokes (two-state folding) and bind to their partners by simple structural complementarity (lock-and-key binding). We now know that a significant fraction of the proteome corresponds to proteins that are intrinsically disordered in their physiological stage. Intrinsically disordered proteins (IDPs) are able to fold upon binding in a process that may provide kinetic advantages and result in complex binding phenomena such as induced-fit and conformational selection. Some IDPs morph upon binding to structurally diverse partners, and others exhibit sophisticated allosteric behavior.
However, there is no quantitative understanding of the molecular mechanisms that lead to the striking conformational features of IDPs. The delicate interplay between folding and binding that must take place in these proteins, and the relationship between folding mechanism and complex binding behavior, remain unanswered questions as of today. There is even no mechanistic information on how the binding of an IDP to a partner influences its binding to other partners, or of how conformational disorder is related to allosteric binding behavior.
In this regard, our group has recently found a direct link between our previous research on fast, non-cooperative (downhill) folding and the marginal stability of the native fold that leads to intrinsically disordered proteins (Campos COSB 2016). This connection explains the structural malleability and complex binding modes found on IDPs in terms of an underlying downhill folding mechanism, which allows the protein to operate as a conformational rheostat. In contrast to the binary patterns of conventional molecular switches, the idea is that conformational rheostats will be able to produce analogical signals, thus opening a new realm of possibilities for the fine control of protein function.
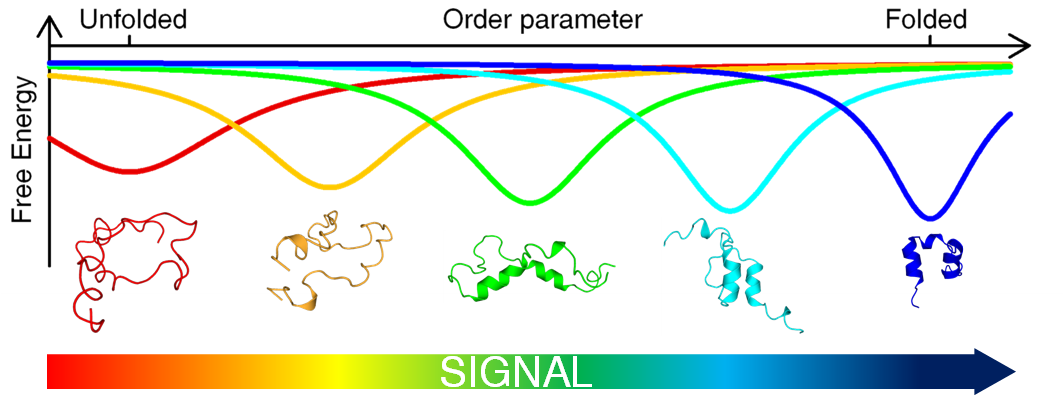
We are very interested in examining the validity of this hypothesis and investigating the roles that conformational rheostats may play in fundamental processes in Biology, most particularly in complex protein-protein interaction networks.
Our current efforts focus on two specific systems:
1. Multienzymatic complexes that catalyze the oxidative decarboxylation of 2-oxo-acids
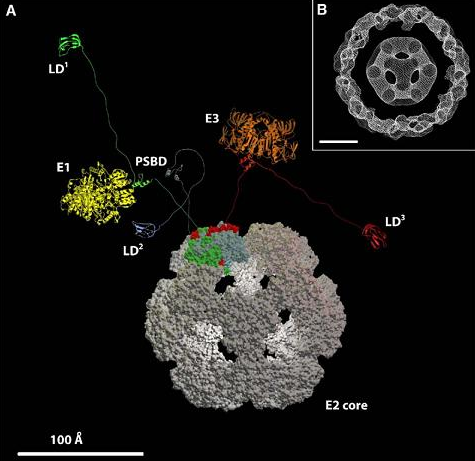
The multi-enzyme complex that we are interested on is composed of a highly dynamic agglomerate of multiple copies of three different enzymes (subunits E1, E2 and E3) which together catalyze the four step reaction. In this highly dynamic protein-protein complex there is one small (~40 residues) central domain of E2 termed peripheral subunit binding domain (PSBD) that plays a key role in the whole process. This role consists of alternately binding to E1 and E3 to coordinate the action of the three enzymes to attain high efficiency. We have discovered that PSBD fold downhill and exhibit significant structural flexibility at physiological conditions, properties that seem to be essential for achieving coordinate sequential binding to E1 and E3. We are now investigating the interplay of interactions between PSBD and E1 or E3 using single-molecule fluorescence and NMR experiments together with computer simulations.
2. The recruitment of multiple proteins to form the transcription machinery.
The CREB binding protein is a multidomain transcriptional coactivator that recruits the basal transcription machinery to the promoter during the early stages of gene expression. Within this protein, it is the 50-residue long nuclear coactivator binding domain (NCBD) that is in charge of binding to the many different proteins involved in transcription activation such as the SRC family, p53, p73, IRF3, and IRF7. We are interested in NCBD because it exhibits the conformational signatures that we expect for a conformational rheostat. In its free state NCBD has great conformational flexibility together with large amounts of alpha-helical structure, which led to its classification as a “structured” IDP. However, in stabilizing conditions (e.g. upon addition of osmolytes) NCBD folds up into a flexible three helix-bundle structure and binds to multiple partners adopting different structures in complex with them, indicating that is structurally malleable by the binding free energy provided by the partner. NCBD has also been classified as a one-state downhill folder based on the multivariate quantitative analysis of experimental data and computer simulations. We are very interested in characterizing the conformational properties of NCBD and investigate the interplay between binding to its various partners.