RESEARCH
Mechanisms of Protein Folding
The protein folding problem has been the major research focus of our group for many years. Over these years we have made multiple seminal contributions, including: the experimental identification of downhill folding (Garcia Mira Science 2002); first measurement of the nanosecond dynamics of hydrophobic collapse (Sadqi PNAS 2003), the identification of scaling effects in folding rates (Naganathan & Munoz JACS 2005), unfolding rates and protein stability (De Sancho JACS 2008); development of analytical methods to determine the thermodynamic barriers to protein folding from differential scanning calorimetry (Munoz PNAS 2004, Naganathan et al JACS 2005, Naganathan PCCP 2011); the experimental characterization of primordial protein folding (Sadqi PNAS 2009); the identification of universal rules defining the energetics of folding transition states (Naganathan PNAS 2010); the development of NMR based methods to map out folding interactions networks (Sadqi Nature 2006, Sborgi JACS 2015); the development of microsecond-resolution single-molecule fluorescence methods to monitor protein folding (Campos Nature Methods 2011, Ramanathan J. Phys. Chem. 2015, Wang Methods in Enzymology 2016); the observation of one-state downhill folding using single-molecule fluorescence detection (Liu PNAS 2012); and the detection of multiple heterogeneous mechanical unfolding pathways on an otherwise two-state folding protein (Schoenfelder Nature Communications 2016).
One of our current interests in this area involve the application of NMR methods to measure the folding interaction networks of a catalogue of folding archetypes, which are single-domain proteins with elementary folding topologies, fast folding kinetics and marginally cooperative unfolding processes. Here we intend to address the question of what is the relationship between native fold and folding mechanism. We are combining our custom NMR experiments with computer simulations to obtain complementary mechanistic information and to use the experimental data as benchmarks for simulations. We are also developing new analytical methods to better understand the connections between chemical shifts (the key NMR parameter that provides folding information at the atomic level) and the broad ensembles of protein structures that are observed in folding experiments and are also the native state of downhill and intrinsically disordered proteins.
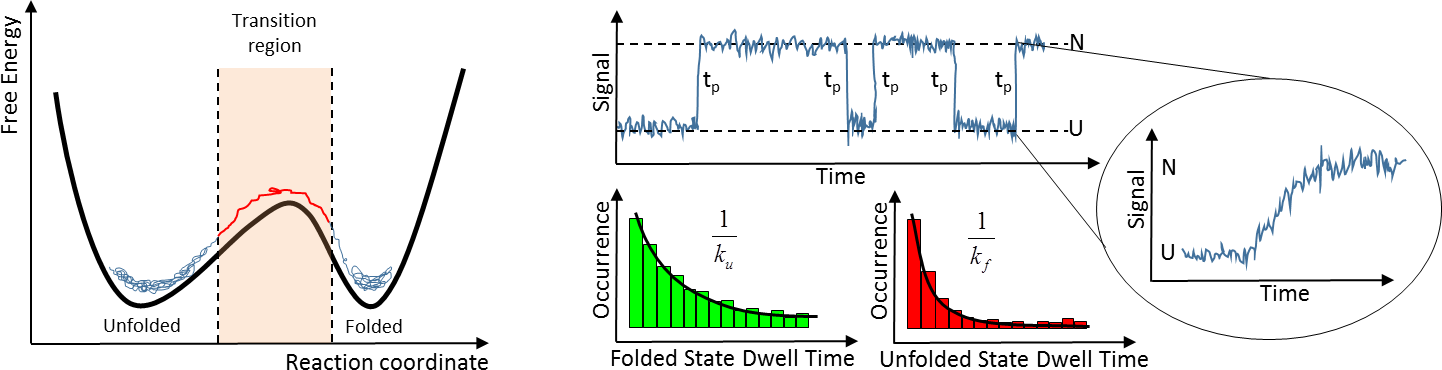
Another major thrust of our current research in protein folding is to be able to resolve experimentally the transition paths that a protein traverses while is folding or unfolding. The transition paths are the reactive segments of a folding trajectory in which the protein leaves one of its ground states (native or unfolded) to traverse the (un)folding free energy barrier. Transition paths are very interesting because they correspond exactly with the process of structure acquisition (or structural melting) and thus hold the keys to folding mechanisms. Transition paths are stochastic processes of individual molecules and thus being able to resolve them requires the application of single-molecule spectroscopic methods that have enough time-resolution (sub-microsecond). We are actively working in developing such methods using both single-molecule fluorescence microscopy or force spectroscopy, and would like to apply them to monitor the folding transition paths of the same catalogue of folding archetypes that we are studying by NMR.